Publications
A near-complete list of publications is available from my Google Scholar page.
Selected manuscripts
Selected manuscripts illustrating past and current work, all with either a first or senior authorship from the Purcell lab (but often conducted with many collaborators or within research consortia):
The Interplay Between Sleep and Mental Health: A Genetic Perspective
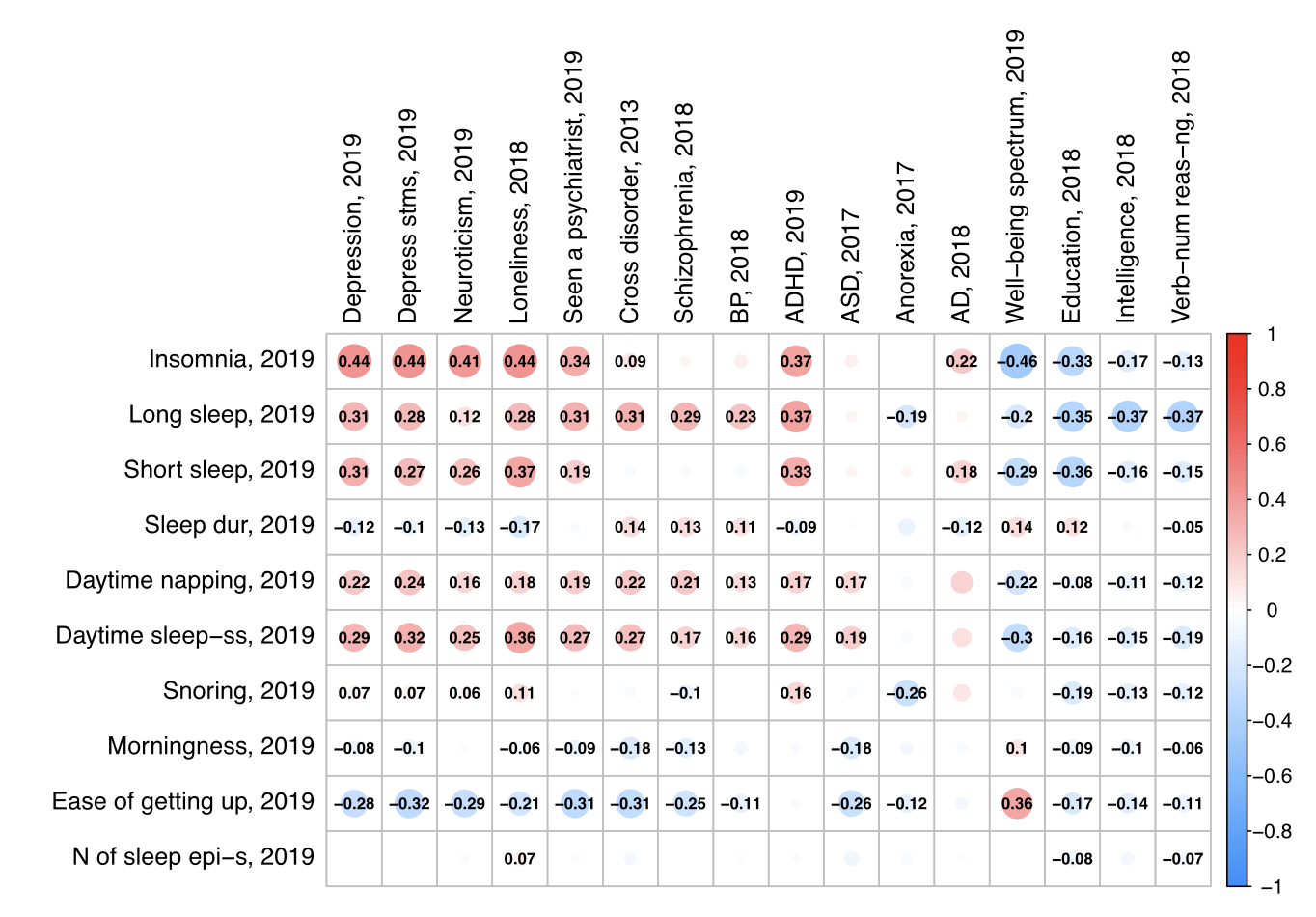
Kozhemiako et al (Biological Psychiatry, 2025) [PDF]
Here, we summarize recent human quantitative and molecular genetic studies on sleep and psychiatric disease, including twin and genome-wide association studies.
Leveraging clinical sleep data across multiple pediatric cohorts for insights into neurodevelopment: the Retrospective Analysis of Sleep in Pediatric (RASP) cohorts study
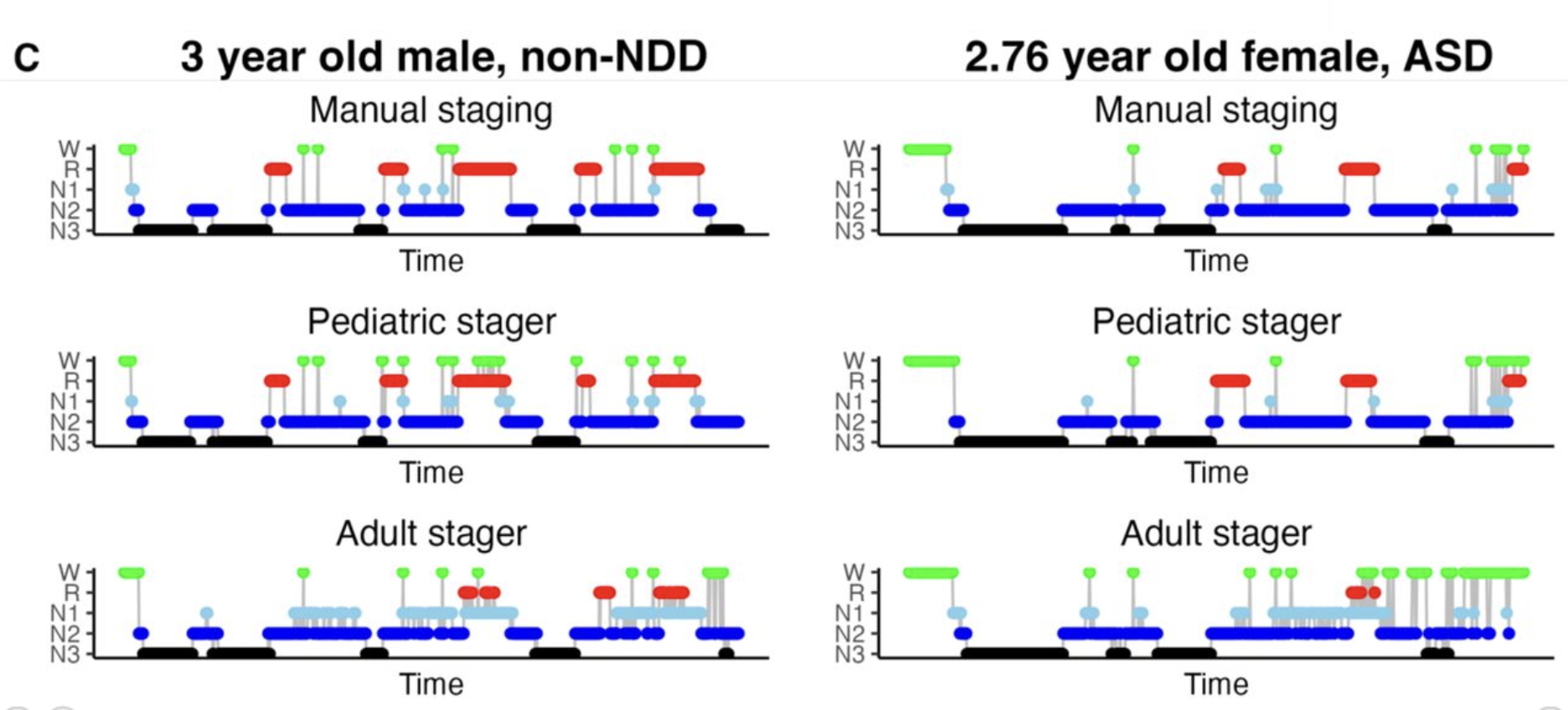
Gong et al (SLEEP, 2026) [PDF]
Here we characterized a retrospective collection of 1527 clinical pediatric overnight PSGs. As well as developing an automated stager trained on independent pediatric sleep data, which yielded better performance compared to a generic stager trained primarily on adults, we built a model to predict biological age from pediatric sleep EEG metrics, which was sensitive to neurodevelopmental delay (NDD) diagnosis.
Mapping the relationships between structural brain MRI characteristics and sleep electroencephalography patterns: the Multi-Ethnic Study of atherosclerosis
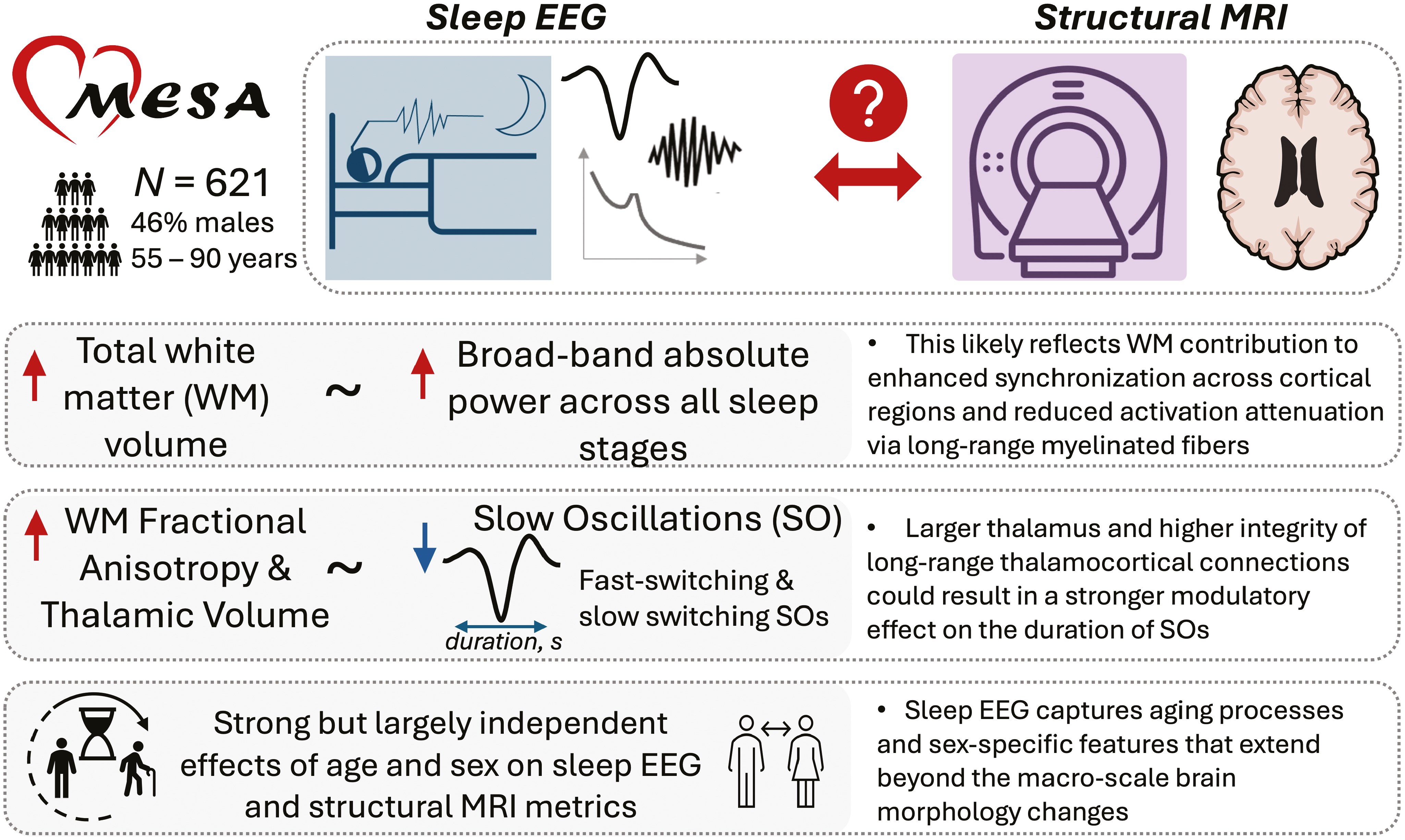
Kozhemiako et al (2025, SLEEP ) [PDF]
We analyzed whole-night sleep electroencephalography (EEG) and brain structural MRI data from a subset of 621 individuals in the MESA study to explore the relationship between brain structure and sleep EEG properties.
Big data approaches for novel mechanistic insights on sleep and circadian rhythms: a workshop summary

Bazier et al (SLEEP, 2025) [PDF]
This report summarizes a workshop, the goals of which were to establish a comprehensive understanding of the current state of sleep and circadian rhythm disorders research and to identify opportunities to advance the field by using approaches based on artificial intelligence and machine learning.
A spectrum of altered NREM sleep in schizophrenia
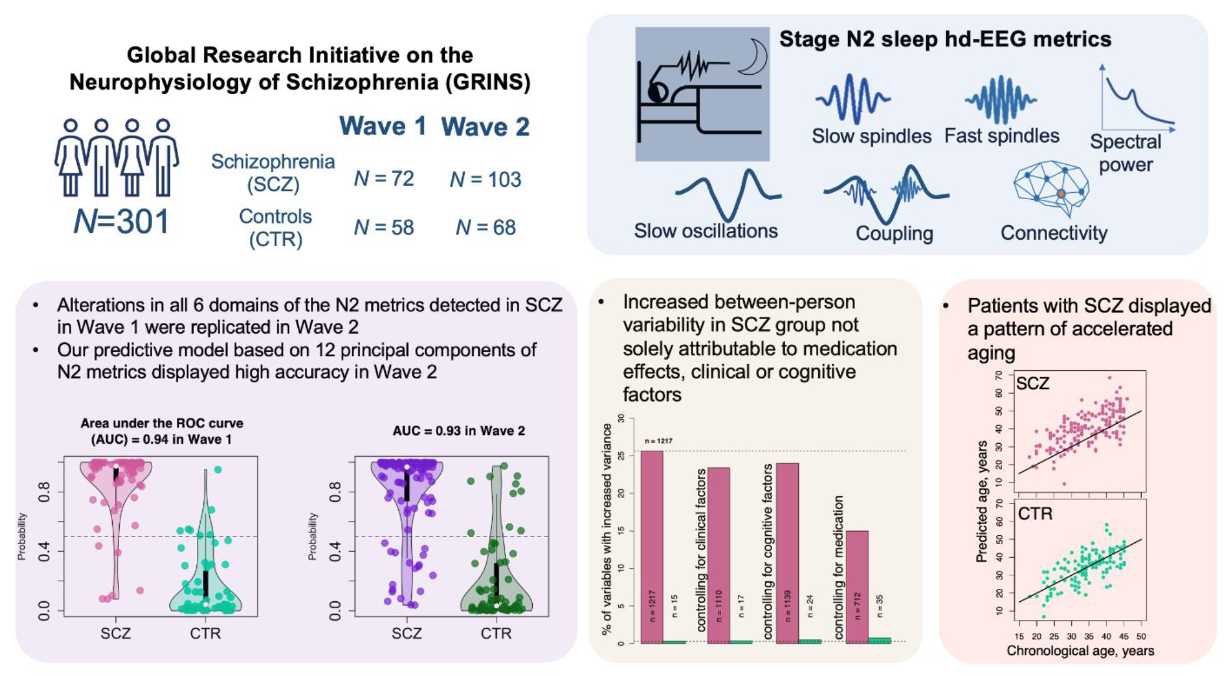
Kozhemiako et al. (SLEEP, 2025) [ PDF ]
Second-wave GRINS results demonstrate a spectrum of objectively measurable N2 sleep deficits in schizophrenia, with implications for etiologic heterogeneity and potential iatrogenic medication effects.
The National Sleep Research Resource: making data findable, accessible, interoperable, reusable and promoting sleep science
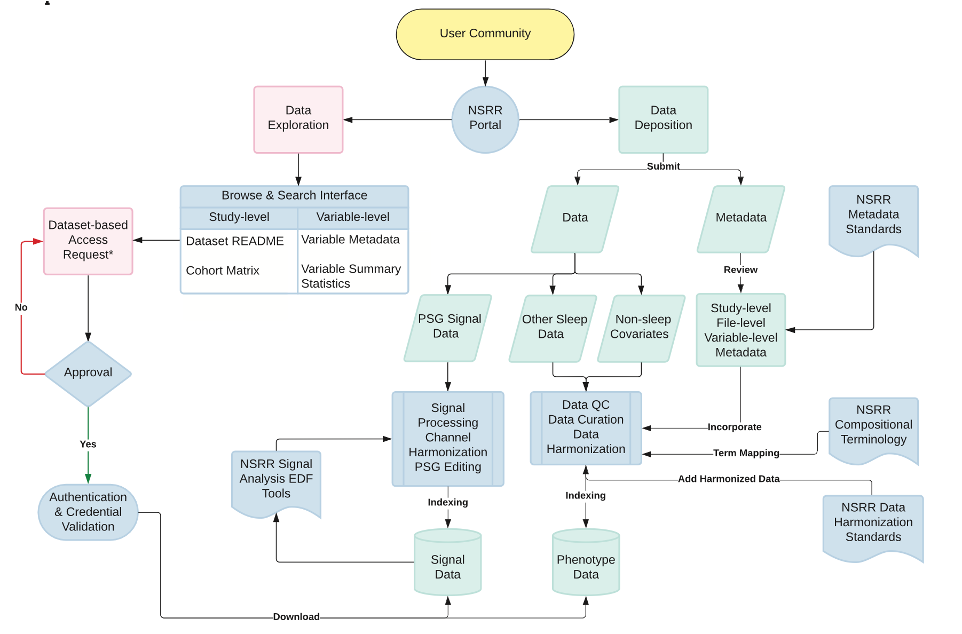
Zhang et al. (SLEEP, 2023) [PDF]
This paper presents a comprehensive overview of the National Sleep Research Resource (NSRR), a National Heart, Lung, and Blood Institute–supported repository developed to share data from clinical studies focused on the evaluation of sleep disorders.
Electroencephalographic Microstates During Sleep and Wake in Schizophrenia
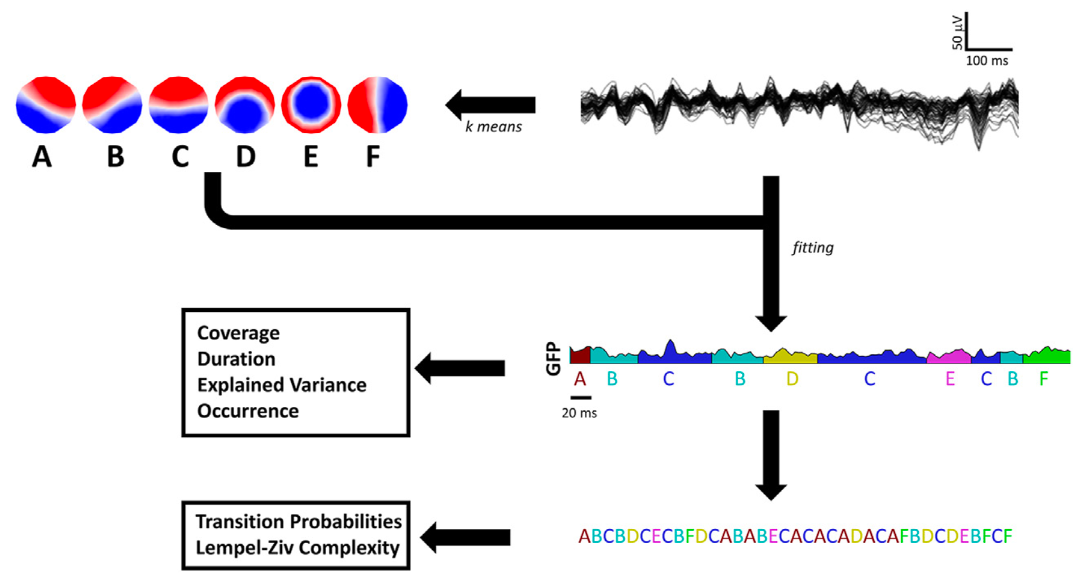
Murphy et al. (Biological Psychiatry GOS, 2024) [PDF]
We analyzed high-density EEG sleep data from 114 patients with schizophrenia and 79 control participants to characterize microstates during sleep and wake. Results reveal behavioral state–dependent cortical dysconnectivity in schizophrenia, largely independent of traditional sleep EEG markers such as reduced spindles.
Mapping neurodevelopment with sleep macro- and micro-architecture across multiple pediatric populations
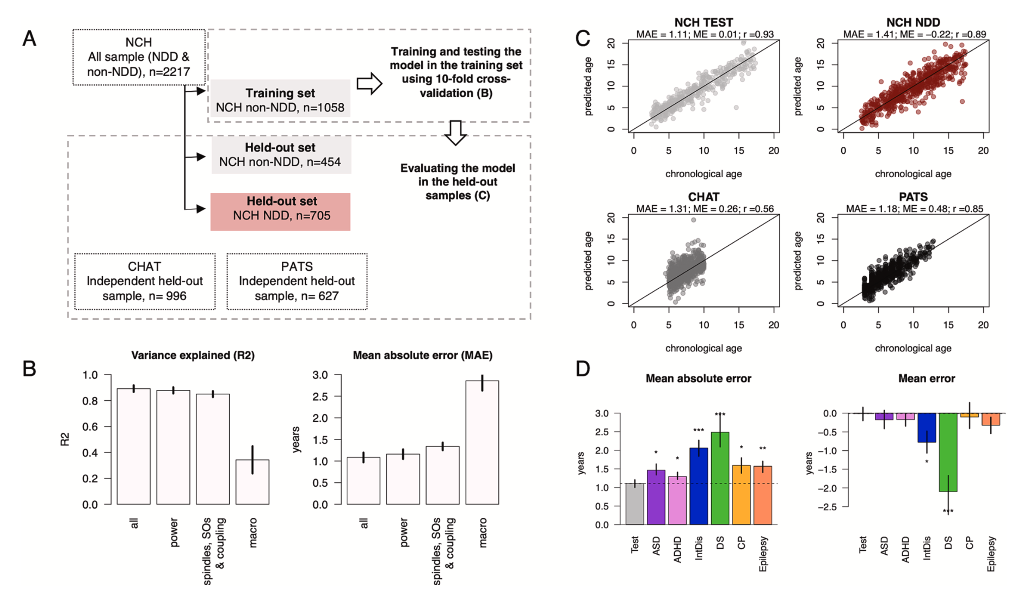
Kozhemiako et al. (NeuroImage: Clinical, 2023) [PDF]
To build and evaluate a sleep-based, quantitative metric of brain maturation, we used PSG data from large research and clinical samples spanning childhood and adolescence (N = 4,013, aged 2.5 to 17.5 years). Our results indicate that sleep architecture offers a sensitive window for characterizing brain maturation, suggesting the potential for scalable, objective sleep-based biomarkers to measure neurodevelopment.
A Potential Source of Bias in Group-Level EEG Microstate Analysis
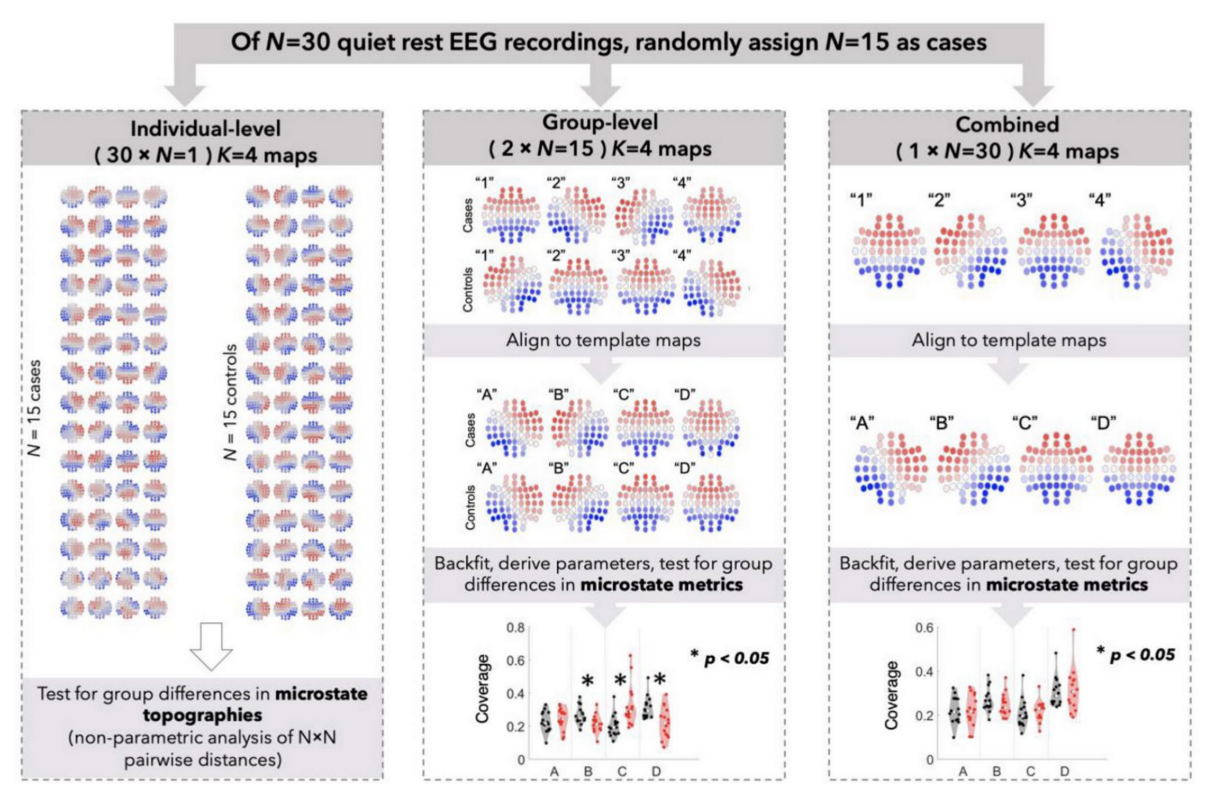
Murphy et al. (Brain Topography, 2023) [PDF]
Microstate analysis is a promising technique for analyzing high-density electroencephalographic data, but there are multiple questions about methodological best practices. Our results suggest that even subtle chance differences in microstate topography can have profound effects on derived microstate metrics and that future studies using microstate analysis should take steps to mitigate this large source of error.
Amyloid beta-independent sleep markers associated with early regional tau burden and cortical thinning
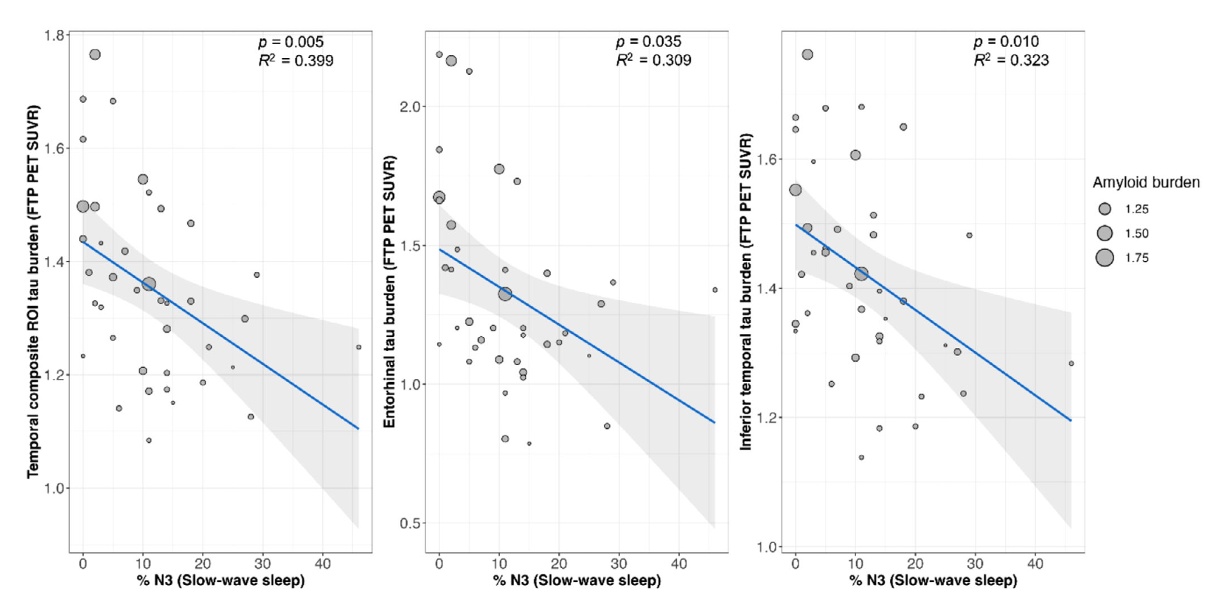
Stankeviciute et al. (Alzheimer’s & Dementia: Amsterdam, 2023) [PDF]
In cognitively unimpaired older adults, lower SWS and higher N1 sleep were associated with higher tau burden and lower cortical thickness in brain regions associated with early tau deposition and vulnerable to AD-related neurodegeneration through mechanisms dissociable from amyloid deposition.
Sources of Variation in the Spectral Slope of the Sleep EEG
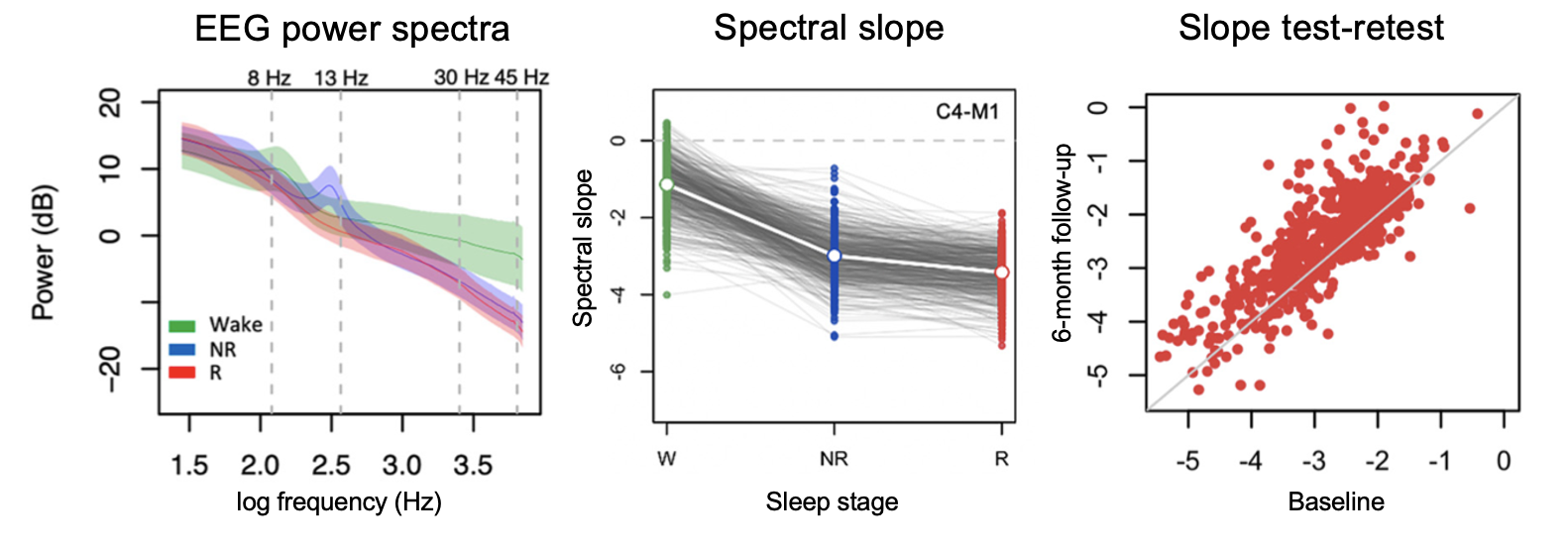
Kozhemiako et al. (eNeuro, 2022) [PDF]
In over 10,000 NSRR PSGs we validate the EEG spectral slope as a neurobiological marker of cortical arousal across wake, NREM and REM sleep, and report sources of intra-individual and inter-individual variability, including changes across the lifespan.
Non-rapid eye movement sleep and wake neurophysiology in schizophrenia
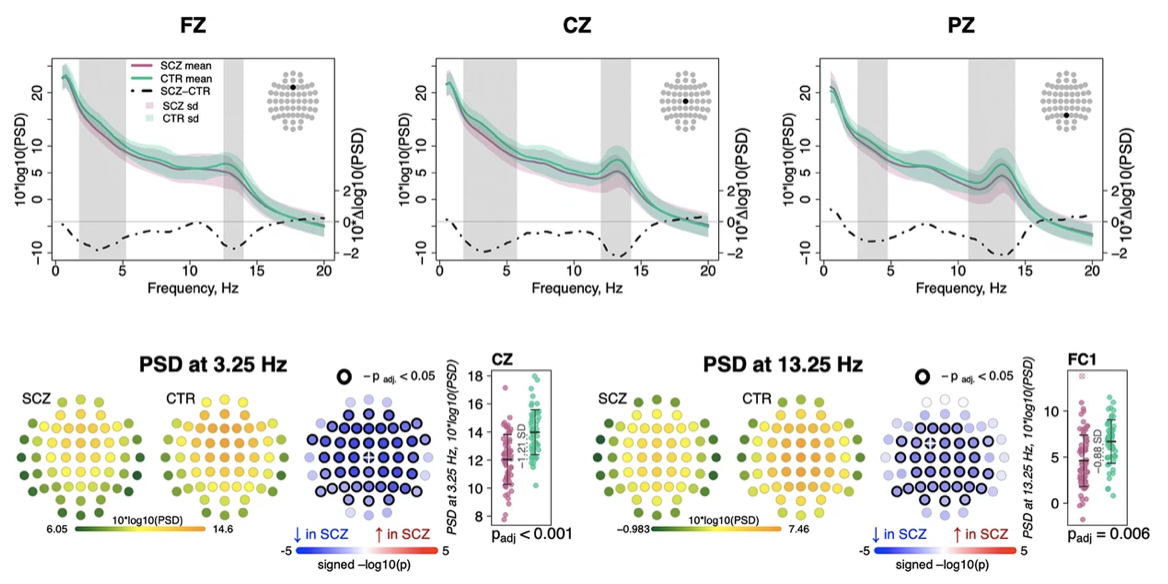
Kozhemiako et al. (eLife, 2022) [PDF]
In a collaboration with the Broad Institute and others, we characterized NREM neurophysiological deficits in schizophrenia.
Macro and micro sleep architecture and cognitive performance in older adults

Djonlagic et al. (Nature Human Behaviour, 2021) [PDF]
This study pointed to multiple facets of sleep neurophysiology that track coherently with underlying, age-dependent determinants of cognitive and physical health trajectories in older adults.
Characterizing sleep spindles in 11,630 individuals from the National Sleep Research Resource
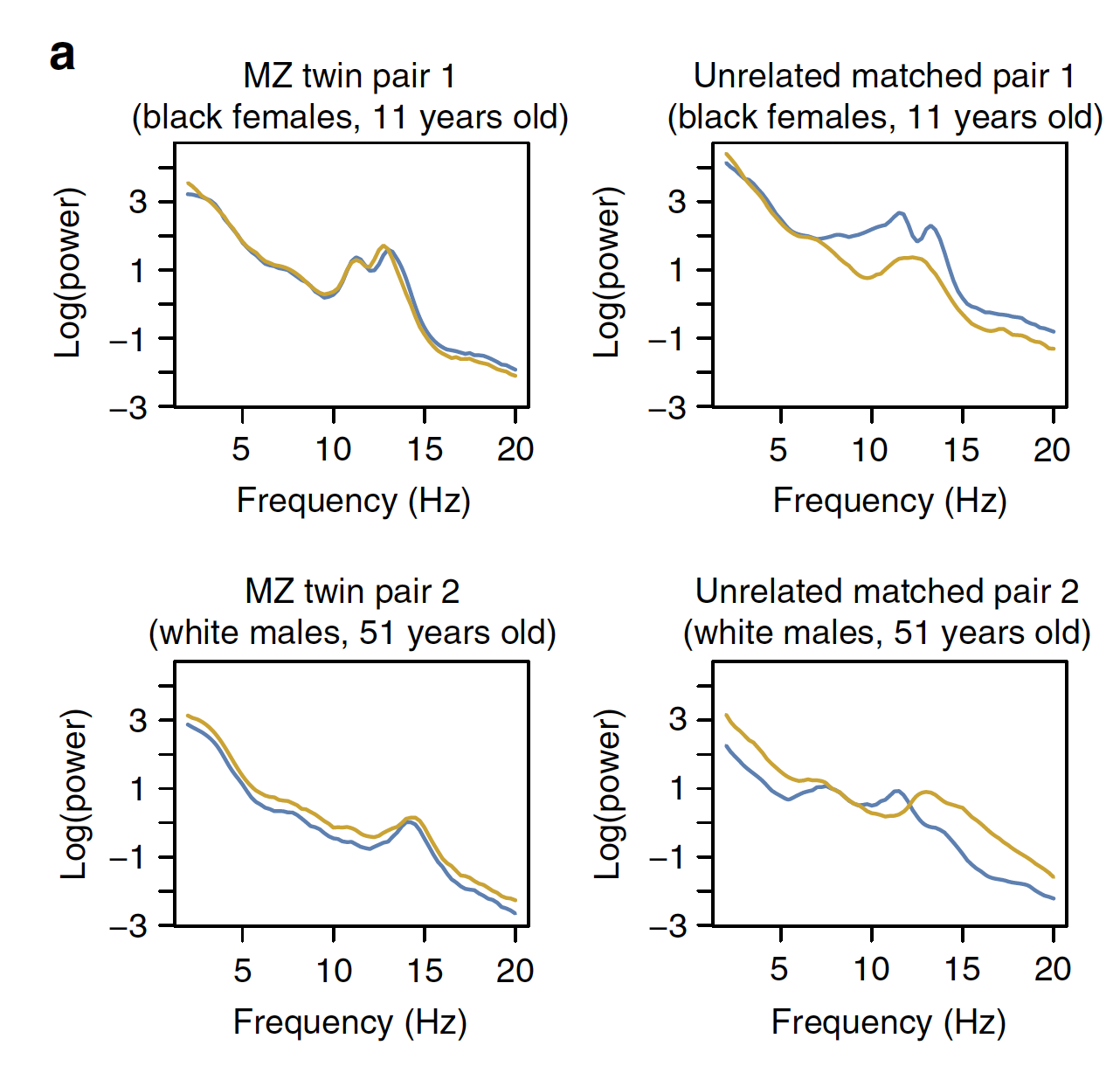
Purcell et al. (Nature Communications, 2017) [PDF]
Here we characterize the considerable variation, both between and within individuals, in sleep spindle activity, demonstrating both developmental trajectories and heritable influence.
A polygenic burden of rare disruptive mutations in schizophrenia

Purcell et al. (Nature, 2014) [PDF]
Exome sequencing over 5,000 Swedish individuals to map schizophrenia alleles, genes and networks. Summary statistics are available here.
De novo mutations in schizophrenia implicate synaptic networks
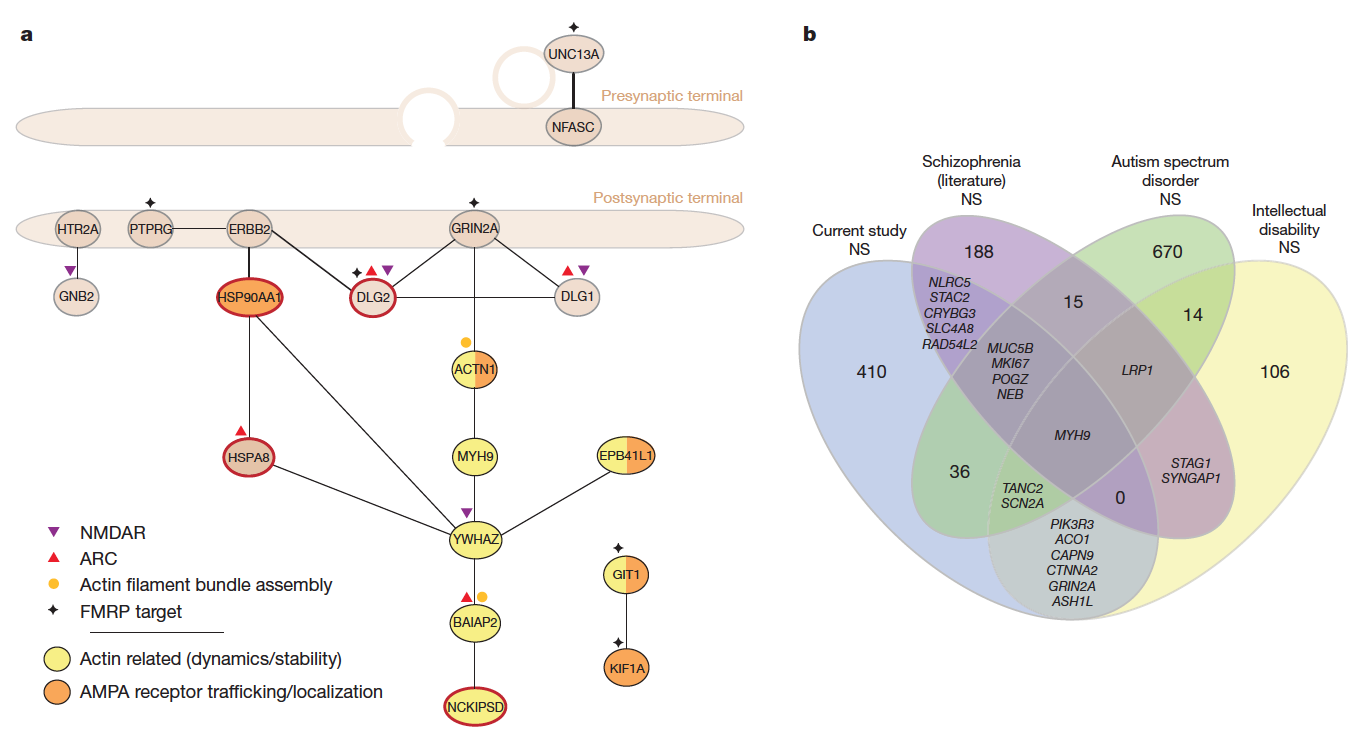
Fromer et al. (Nature, 2014) [PubMed | PDF]
Using next-generation exome sequencing in over 600 families to determine the role of newly arising mutations in schizophrenia. Summary statistics are available here.
PLINK: a tool set for whole-genome association and population-based linkage analyses

Purcell et al. (American Journal of Human Genetics, 2007) [PubMed | PDF]
This describes PLINK, a widely used software package for genome-wide association studies. We also introduced a novel approach to genome mapping based on identity-by-descent (IBD) information between very distantly related individuals.
Common polygenic variation contributes to risk of schizophrenia and bipolar disorder
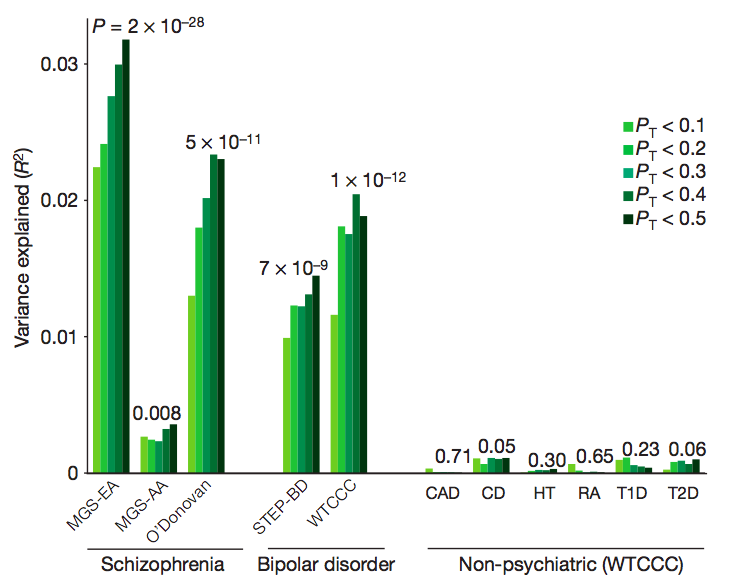
International Schizophrenia Consortium (Nature, 2009) [ PDF]
One of the first large-scale genome-wide association studies of schizophrenia, here we made the observation that many common variants of very small effect appear to contribute to the heritability of schizophrenia. Most of these risk alleles are individually unlikely to be detectable, at least at currently feasible sample sizes.
Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth (XHMM)
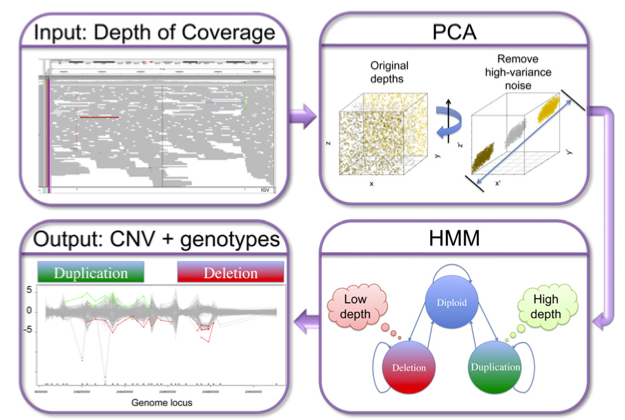
Fromer et al. (American Journal of Human Genetics, 2012) [PDF]
Reliably calling copy number variation from targeted sequencing data can be challenging. Here we present and evaluate a robust pipeline (XHMM) that overcomes many of the challenges that arise when trying to infer ploidy from sequencing read depth in exome sequencing studies.
Rare chromosomal deletions and duplications increase risk of schizophrenia

International Schizophrenia Consortium (Nature, 2008) [PDF]
Using genome-wide single nucleotide polymorphism arrays, we demonstrated an increased genome-wide burden of rare deletions and duplications (copy number variants, CNV) in patients with schizophrenia compared to controls. We also identified specific genomic loci at which CNVs pile up, namely 22q11, 15q13 and 1q21. [commentaries here and here]
Power calculations and significance testing in genetic studies
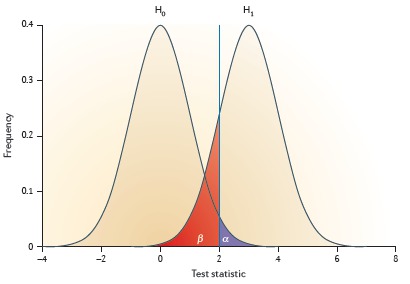
Sham & Purcell (Nature Reviews Genetics, 2014) [PDF]
An introduction and review of the issues around significance testing and power calculation in large-scale genetic studies.
Pleiotropy in complex traits: challenges and strategies
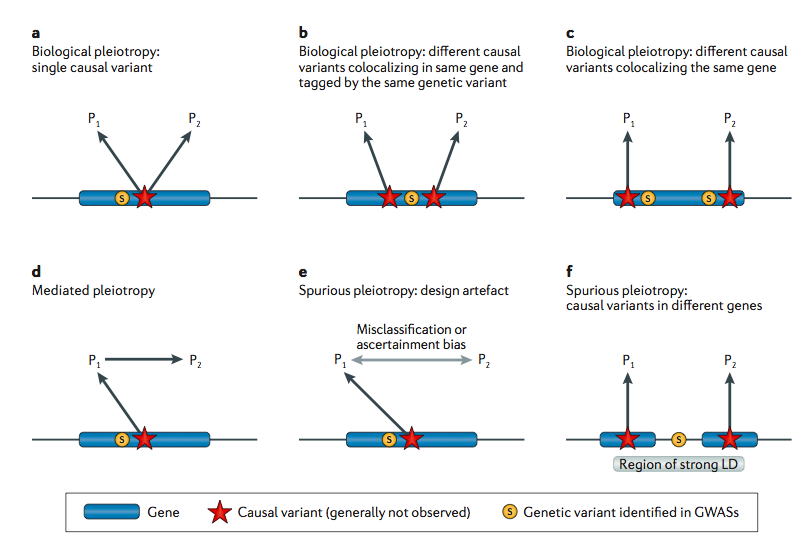
Solovieff et al. (Nature Reviews Genetics, 2013) [PDF]
This Review considers approaches and interpretations of the often “one-to-many” nature of genotype-to-phenotype associations in complex disease. Nadia Solovieff was a postdoctoral researcher in the Purcell lab, jointly with Dr. Jordan Smoller’s group.
Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder
Ferreira et al. (Nature Genetics, 2008) [PDF]
This manuscript was the first meta-analysis of genome-wide single nucleotide polymorphism (SNP) data across studies of bipolar disorder. We implicated several genomic regions, including a role for calcium ion channel genes in this disease. [commentary]
Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4
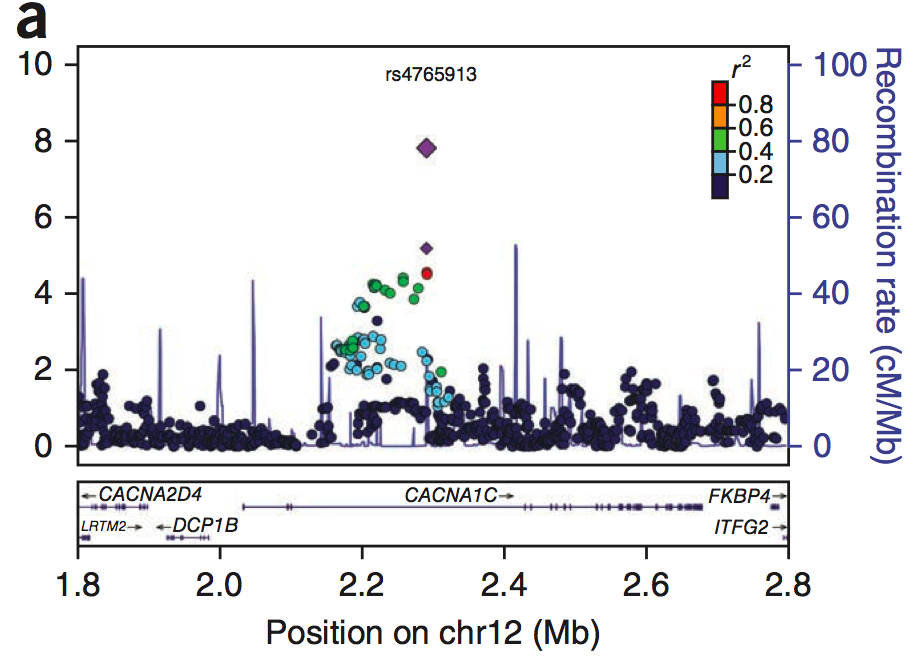
Sklar et al. (Nature Genetics, 2012) [PDF]
A later combined analysis of over 15,000 individuals, performed within the context of the PGC, provided further support for calcium channel and other genes.
Variance components models for gene-environment interaction in twin analysis
Purcell (Twin Research, 2002) [PDF]
A relatively old publication, included here to indicate some of our earlier work in behavioral genetics and twin studies. This manuscript provided a comprehensive methodological treatment for modeling how measured environmental variables can modify the relative impact of genetic influences for quantitative traits.